Learning CenterAccess additional learning opportunities. |
Summer School
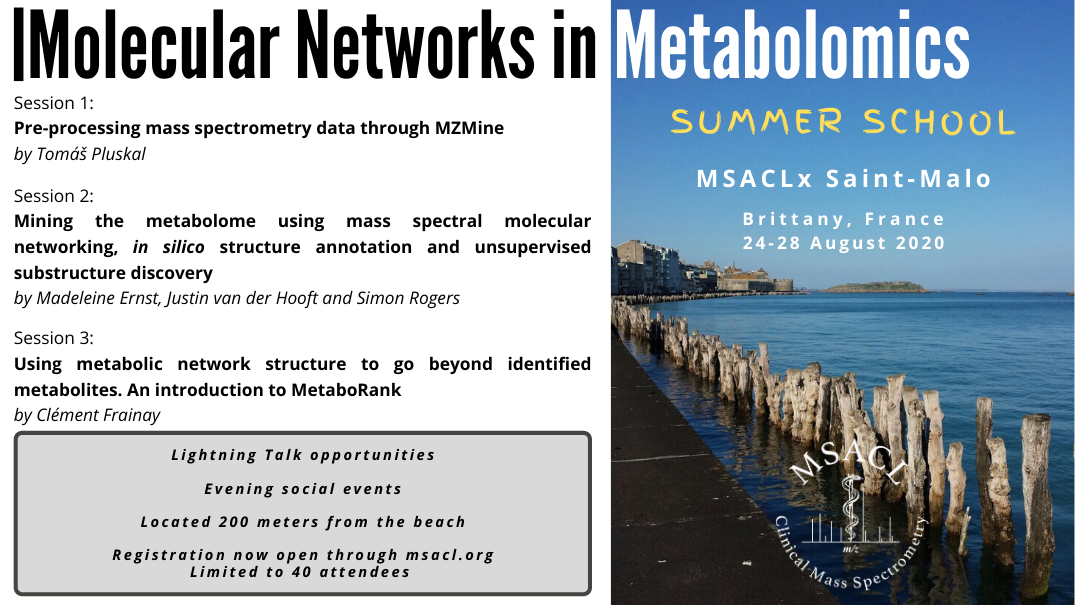
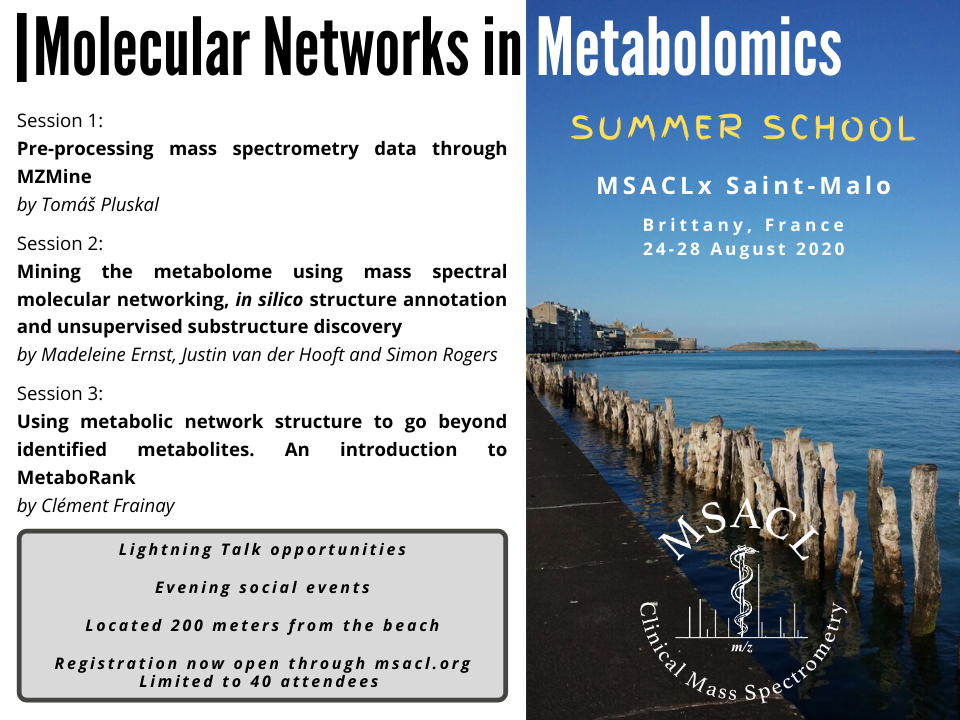
1st MSACL Summer School ProgrammeMonday 24th to Friday 28th of August 2020 Topic: Molecular Networks in Metabolomics Capacity : 40 students
Ethic Etapes Auberge de Jeunesse |
On-site Organizer Marine Letertre m.letertre16@gmail.com |
|
Course Instructors Registration Fee and Details of Coverage Preliminary Schedule Session 1 | Evening Workshop | Session 2 | Session 3 How to Get There Where to Eat Sponsorship Promotional Media
|
CANCELLED
Cancellation Policy |
Metabolomics, defined as the measurement of all small molecules detected in a given biological sample, has gained a great importance within the field of clinical mass spectrometry. The metabolome is directly linked to key factors influencing health and disease, such as genetics, the environment, and the microbiome and thus offers a unique insight into metabolic mechanisms mediating health and disease.
However, a key challenge remains in the high-throughput metabolite identification indispensable for biological interpretation. In recent years, an increasing number of computational metabolomics tools have started to embrace this challenge.
During this four-day summer school, students will learn how to preprocess liquid chromatography tandem-mass spectrometry (LC-MS/MS) based metabolomics data to retrieve quantitative information using MZmine. Furthermore, the students will learn how to retrieve chemical structure information using the Global Natural Products Social Molecular Networking (GNPS) platform, in silico structure annotation and unsupervised substructure discovery using MS2LDA. Finally, the network-based algorithm MetaboRank will be taught to facilitate metabolite annotation in the context of biochemical pathways.
Course Instructors | ||||
 |
 |
 |
 |
 |
| Tomáš Pluskal, PhD Institute of Organic Chemistry and Biochemistry, Czech Academy of Sciences, Prague, Czech Republic |
Madeleine Ernst, PhD Center for Newborn Screening, Department of Congenital Disorders, Statens Serum Institut, Copenhagen, Denmark |
Justin van der Hooft, PhD Bioinformatics Group, Plant Sciences Group, Wageningen University, Wageningen, Netherlands |
Simon Rogers, PhD School of Computing Science, University of Glasgow, Glasgow, United Kingdom |
Clément Frainay, PhD INRAe UMR1331 Toxalim Research Centre in Food Toxicology, University of Toulouse, Toulouse, France |
Registration Fee and Details of Coverage
Join this 4-day summer training workshop for 500 EUR.
Registration Includes:
- A shared double room at the Auberge de Jeunesse in Saint Malo for arrival on Monday August 24 and departure on Friday August 28.
- Breakfast at the Auberge on Tuesday, Wednesday, Thursday and Friday.
- Lunch at the Auberge on Tuesday, Wednesday, Thursday and Friday (Friday is brown bag take-away).
⇒ Diet-specific options available during registration or by contacting Marine Letertre. - A Tuesday welcome Dinner catered at the Auberge to include crepes, galettes and beverages.
- A Thursday very special Gala dinner!
- Coffee & Biscuit breaks during the training sessions.
The only meals for which the student will need to cover costs are dinner on Monday, Wednesday & Friday.
Preliminary Schedule
Monday 24th of August:
6 pm: Registration opens : Pick up badge and room key.
Tuesday 25th of August:
7.30 to 8 am: Registration continues : Pick up badge and room key.
8 am: Opening introduction
8.15 am to 12.25 pm: Session 1
12.30 pm: Lunch
2 to 5.30 pm: Session 1
5.45 to 6.30 pm: Lightning talks
6.30 pm: Welcome Drinks and Dinner, Informal typical Brittany dinner (with crêpes and galettes) and Evening Workshop
Wednesday 26th of August:
8 am to 12.25 pm: Session 2
12.30 pm: Lunch
2 to 5.30 pm: Session 2
5.45 to 6.30 pm: Lightning talks
6.30 pm: City tour (45 to 60 minutes walk) from the location of the summer school to the old town by the beach, where drinks and dinner on our own can be enjoyed.
Thursday 27th of August:
8 am to 12.25 pm: Session 2
12.30 pm: Lunch
2 to 5 pm: Session 3
5.15 to 5.45 pm: Industrial talks & Lightning talk prize presentation
5.45 pm: Leaving for Gala Dinner
Friday 28th of August:
8 am to 1.00 pm: Session 3
1.00 pm to 1.05 pm: Closing Remarks
1.05 pm: Pick up brown bag lunch at Auberge cantine
Session 1: Preprocessing mass spectrometry data through MZmine
 |
| Tomáš Pluskal, PhD Institute of Organic Chemistry and Biochemistry, Czech Academy of Sciences, Prague, Czech Republic |
Content
The MZmine section will teach basic methods for mass spectrometry data preprocessing, including feature detection, deisotoping, alignment, and gap filling. We will take advantage of the various visualization modules in MZmine to optimize the choice of parameters during these steps. We will also learn how to use various metabolite identification tools, including spectral database searching and machine learning- based prediction methods such as CSI-FingerID. Finally, we will export the results from MZmine for downstream statistical analyses and for molecular networking tools.
Goals and learning points
i) how to preprocess mass spectrometry data using MZmine
ii) how to optimize preprocessing parameters using MZmine visualization modules
iii) how to use metabolite identification tools
iv) how to export MZmine-preprocessed data for further analyses and molecular networks
Evening Workshop: Molecular networks based on t-SNE algorithm : an initiation to MetGem
 |
| David Touboul, PhD Natural Product Chemistry Institute, Paris, France |
Content
Molecular networking (MN) is becoming a standard bioinformatics tool in the metabolomic community. The problem of creating a reliable representation of MS2 spectra data sets can be solved using algorithms developed for dimensionality reduction and pattern recognition purposes, such as t-distributed stochastic neighbor embedding (t-SNE). This multivariate embedding method pays particular attention to local details by using nonlinear outputs to represent the entire data space. Our software, MetGem, allows the parallel investigation of two complementary representations of the raw data set, one based on a classic GNPS-style MN and another based on the t-SNE algorithm. The t- SNE graph preserves the interactions between related groups of spectra, while the MN output allows an unambiguous separation of clusters. Almost all parameters can be tuned in real time, and new networks can be generated within a few seconds for small data sets. With the development of this unified interface (https://metgem.github.io), we fulfilled the need for a dedicated, user-friendly, local software for MS2 comparison and spectral network generation.
Session 2: Mining the metabolome using mass spectral molecular networking, in silico structure annotation and unsupervised substructure discovery
 |
 |
 |
| Madeleine Ernst, PhD Center for Newborn Screening, Department of Congenital Disorders, Statens Serum Institut, Copenhagen, Denmark |
Justin van der Hooft, PhD Bioinformatics Group, Plant Sciences Group, Wageningen University, Wageningen, Netherlands |
Simon Rogers, PhD School of Computing Science, University of Glasgow, Glasgow, United Kingdom |
Content
GNPS, in silico structure annotation and MS2LDA provide solutions towards enhanced metabolome mining and structural annotation as well as providing a framework for data sharing. This workshop will enhance the toolkit of the modern clinical metabolomics researcher in maximizing structural information retrieved from a metabolomics experiment as well as highlighting the importance and benefits of data sharing.
Goals and learning points
We will introduce automated tandem mass spectrometry-based dereplication using a combination of the Global Natural Products Social Molecular Networking (GNPS) platform, in silico structure annotation (NAP, SIRIUS+CSI:FingerID followed by MolNetEnhancer) and unsupervised substructure discovery through MS2LDA.
Participants will learn:
i) how to submit MZmine preprocessed mass spectrometry data to GNPS
ii) how to run a feature-based mass spectral molecular networking job on the GNPS interface and which parameters are important to consider
iii) how to browse through the results on the GNPS interface
iv) how to run in silico structure prediction through NAP and SIRIUS+CSI:FingerID
v) how to browse through in silico structure results on the GNPS interface
vi) how to run an MS2LDA job on the GNPS interface and which parameters are important to consider
vii) how to browse through the MS2LDA results on the GNPS interface
viii) how to integrate chemical structural information from mass spectral molecular networking, in silico structure annotation and unsupervised substructure discovery through the MolNetEnhancer workflow
to
highlight broad chemical classes and substructural differences
Format
This will be a hands-on session where participants submit example data to GNPS, perform in silico structure annotation and MS2LDA substructure discovery. Theoretical background as well as practical examples for integration and data interpretation of all platforms will be provided. The session will be interactive, with plenty of room for questions and feedback from the participants. Clinically relevant example data will be provided, however participants are also encouraged to bring their own data.
Session 3: Using metabolic network structure to go beyond identified metabolites. An introduction to MetaboRank
 |
| Clément Frainay, PhD INRAe UMR1331 Toxalim Research Centre in Food Toxicology, University of Toulouse, Toulouse, France |
Content
Knowledge about the metabolic capacity of an organism can be intuitively represented as a network of compounds, connected by the biochemical reactions that consume and produce them, i.e. a Genome-Scale Metabolic Networks (GSMN). The structure of this network can provide valuable information for interpreting metabolomic results, by putting them in the context of the whole system they're part of. This session is aimed for a gentle introduction to network science and graph theory, presenting concepts and analysis most relevant to the metabolomician. It will highlight the common pitfalls and challenges specific to the application to GSMN. A practical session will be dedicated to the information extraction and visual exploration of metabolic networks in MetExplore, and the use of MetaboRank to find new metabolite candidates to investigate.
Goals and learning points
1.1) Understanding GSMN: how they are built, their use and their limitations
1.2) Understanding the basic concepts behind network science
1.3) Interpreting distances and topological measures in metabolic network
2.1) How to browse information about metabolism in MetExplore
2.2) How to map metabolomics data onto a metabolic network in MetExplore
2.3) How to visualise and explore networks with MetExploreViz
2.4) How to go beyond a signature using MetaboRank recommender system
Format
The session will start by a theoretical lecture on biological network analysis focused on metabolomic results interpretation. It will be followed by a brief review of existing tools for network analysis, and a hands-on session on MetExplore, a web server dedicated to the analysis and visualisation of GSMN.
How to Get There
Where to Eat
Around the Auberge:
- Restaurant Patrick Varangot, directly at the Auberge from 7.30 to 8 pm
- Portobello, 9 minutes walk, garden
- Les Charmettes, €€, 5 minutes walk, beach view
- Le Coude à Coude, 10 minutes walk, french restaurant
- Bar rooftop of the Ambassadeurs Hotel, 15 minutes walk, beach and city view
In the Old town (30 minutes walk by the beach or 15 minutes by bus):
- Les Terroiristes Associés, Wine bar and bistro, €€
- Le Penjab, Indian restaurant
- La Rose Des Vents, Crêperie
- La Trinquette, Bar
- La Belle Epoque, Bar
- Le Chateaubriand, French restaurant
Around Solidor (20 minutes by bus):
- La Cale, Seafood restaurant
- Le Cancalais, Bar Tapas
Sponsorship
Promotional Media
| Slide 16:9 | Slide 4:3 |
|
|